Xiaozhe ("XZ") Ding
Biotech Co-founder (Receptive Bio) | Molecular Engineer
Ph.D. in Bioengineering (minor in CSE), Caltech
B.Sc. in Biological Science (honors), Tsinghua
Building receptor-mediated, therapeutic delivery platforms for the brain and beyond through structure-guided molecular design, fusing AI-powered modeling with wet-lab validation.
Working on: biomolecular engineering (experimental & computational) • drug delivery
Interested in: protein structures • protein design • blood–brain barrier • synthetic biology • molecular biophysics • gene-therapy vectorology • aging
Research | 研究
Please find my up-to-date lists of publications at: Google Scholar / ORCID / ResearchGate
Ph.D. projects
2016 Summer - 2023 Spring
Advisor: Dr. Viviana Gradinaru (Gradinaru Lab )
Ph.D. program in Bioengineering
Division of Biology and Biological Engineering, Caltech
Thesis: Computation-aided protein engineering for targeted therapeutic delivery. Link
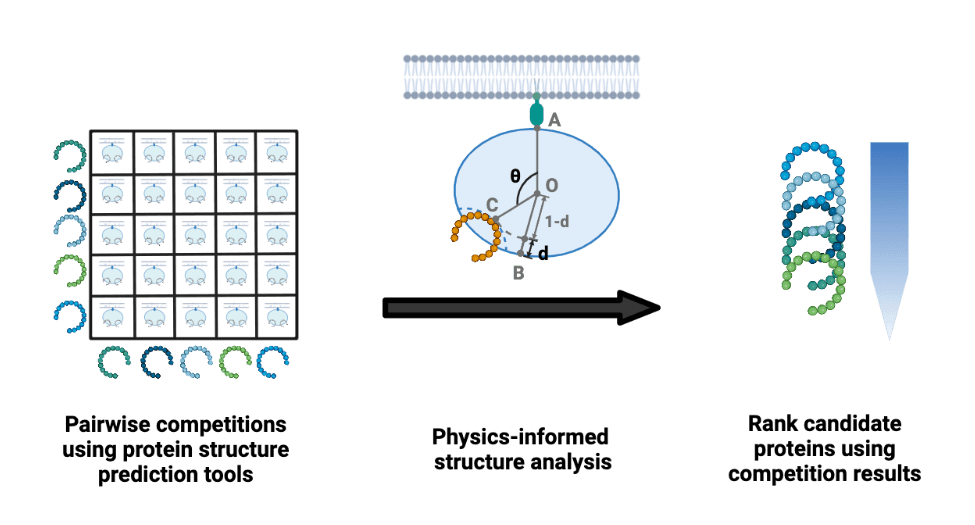
APPRAISE: Rank engineered binders using structure modeling
2021-2023
APPRAISE is a computational method for predicting the receptor binding propensity of engineered proteins based on structural modeling. This tool, leveraging the most recent breakthroughs in computational structure prediction (e.g., AlphaFold2), may help protein engineers evaluate protein/peptide-based therapeutics for targeted drug delivery. You can try APPRAISE in your browser using Google Colab. (bioRxiv Manuscript, GitHub repository)
Related publication:
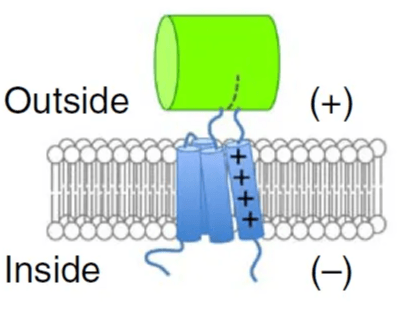
Elucidating the molecular mechanism underlying brain-tropic viral vectors
2018-2023
The brain is protected by a tight blood-brain barrier (BBB) that prevents most macromolecules from accessing the brain. To enable the delivery of genetic therapeutics to the brain, bioengineers have identified a panel of brain-transducing gene delivery vectors based on Adeno-associated virus (AAV), a non-pathogenetic virus that symbioses with primates, using a protein engineering technique called directed evolution. I developed and applied computational pipelines based on AlphaFold to help elucidate how these viral vectors cross the BBB at molecular resolution. These novel mechanisms for brain delivery are helping biomedical scientists to make safer and more effective therapeutics to treat brain diseases.
Related publications:
- Shay, T. F.†*, Sullivan, E. E.†, Ding, X.† (co-first), Chen, X., Kumar, S. R., Goertsen, D., ... & Gradinaru, V.* (2023). Primate-conserved Carbonic Anhydrase IV and murine-restricted Ly6c1 are new targets for crossing the blood-brain barrier. Science Advances. †: Equal contribution. Link News
- Jang, S., Shen, H. K., Ding, X., Miles, T. F., & Gradinaru, V.* (2022). Structural basis of receptor usage by the engineered capsid AAV-PHP. eB. Molecular Therapy-Methods & Clinical Development. Link
Related patents:
- Shay, T. F., Gradinaru V., Ding, X. Compositions and methods for crossing blood brain barrier (US18/177,566). Link
Viral vector capsid engineering
2017-2023
Expand the size of AAV capsids. We aimed to increase the cloning capacity of AAV capsids by expanding the sizes of single particles (3-minute video by Caltech Neuroscience).
Biophysical methods for AAV capsid engineering. I helped develop new biophysical methods and models that facilitated multiple AAV capsid engineering projects.
Related publications:
- Chen, X., Wolfe, D. A., Bindu, D. S., Zhang, M., Taskin, N., Goertsen, D., ... & Gradinaru, V.* (2023). Functional gene delivery to and across brain vasculature of systemic AAVs with endothelial-specific tropism in rodents and broad tropism in primates. bioRxiv. Link
- Seo, J. W., Ingham, E. S., Mahakian, L., Tumbale, S., Wu, B., Aghevlian, S.,Shams S., Baikoghli M., Jain P., Ding X., Goeden N. ... & Ferrara, K. W.* (2020). Positron emission tomography imaging of novel AAV capsids maps rapid brain accumulation. Nature communications. Link
- Ravindra Kumar, S., Miles, T. F., Chen, X., Brown, D., Dobreva, T., Huang, Q., Ding X., Luo Y. ... & Gradinaru, V.* (2020). Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nature methods. Link
Rotation projects

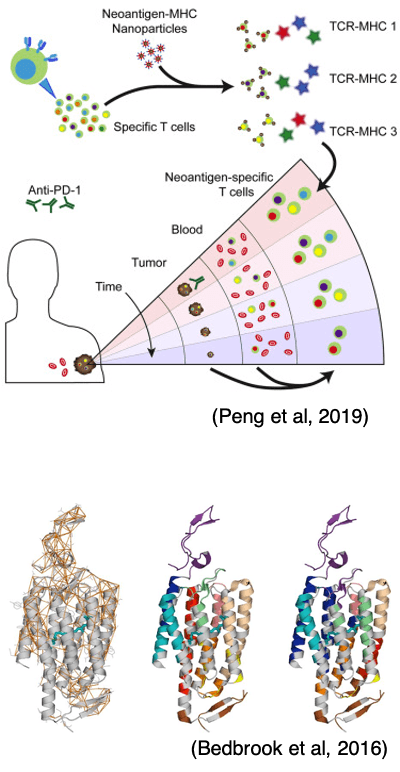
Structure-guided directed evolution of channelrhodopsins
2016 Spring
Advisor: Dr. Frances Arnold (Arnold lab, Nobel Prize 2018)
Co-mentor: Dr. Claire Bedbrook
Division of Chemistry and Chemical Engineering, Caltech
We engineered a genetically encoded tool for optical control of neuronal activity by structure-guided recombination.
Related publication:
- Bedbrook, C.N., Rice, A.J., Yang, K.K., Ding, X., Chen, S., LeProust, E.M., Gradinaru, V. and Arnold, F.H., (2017). Structure-guided SCHEMA recombination generates diverse chimeric channelrhodopsins. Proceedings of the National Academy of Sciences. Link
Technology development for sensitive detection and analysis of neoantigen-specific T cells
2015 Winter
Advisor: Dr. James Heath (Heath lab, now Heath lab @ISB)
Co-mentor: Dr. Songming Peng
Division of Chemistry and Chemical Engineering, Caltech
I supported the development of a technology for the extraction and analysis of neoantigen-specific T cells from cancer patients.
Related publication:
- Peng, S., Zaretsky, J.M., Ng, A.H., Chour, W., Bethune, M.T., Choi, J., Hsu, A., Holman, E., Ding, X., Guo, K. and Kim, J.,..., Baltimore, D., Ribas, A., Heath, J.R. (2019). Sensitive detection and analysis of Neoantigen-specific T cell populations from tumors and blood. Cell reports. Link
Protein engineering for non-invasive neural modulation
2015 Fall
Advisor: Dr. Mikhail Shapiro (Shapiro lab)
Co-mentor: Dr. Jérôme Lacroix & Di Wu
Division of Chemistry and Chemical Engineering, Caltech
We explored a few designs of genetically encoded tools for acoustic modulation of neuronal activity.
Related publication:
- Acknowledged for contributing to initial experiments in Wu, D., Baresch, D., Cook, C., Ma, Z., Duan, M., Malounda, D., ... & Shapiro, M. G. (2023). Biomolecular actuators for genetically selective acoustic manipulation of cells. Science Advances. Link
Undergraduate projects

A ratiometric fluorescent calcium indicator
2014 Spring
Advisor: Dr. Xiaodong Liu (The "X-lab")
Co-mentor: Dr. Yaxiong Yang
Departments of Biomedical Engineering, Tsinghua
Genetically encoded calcium indicators (GECIs) are engineered proteins whose fluorescence intensity indicates calcium concentration, but currently available GECIs have limitations in either dynamic range or accuracy. To overcome the limitations, I designed two novel GECIs, one of which was to fuse two existing GECIs: RCaMP and Inverse-Pericam. The red fluorescence of RCaMP rises when calcium concentration increases, while Inverse-Pericam gives a negative calcium response in yellow fluorescence, and the ratio between red fluorescence and yellow fluorescence is used as the readout for calcium concentration. By doing so, I expected that this new sensor could have lower background noise and a broader dynamic range. Co-expression experiments indeed showed some promising results. Unfortunately, I could not overcome the difficulty in expressing functional fusion protein in multiple trials before I graduated.
This work received "Best Research Award" in the annual retreat of Tsinghua Talented Program (Link to news). The training was sponsored by Tsinghua Talented Program ("Xuetangban" Program).Intro reviews on chemical biology topics
2014 - 2015
Advisors: Dr. Lei Liu, Dr. Yiming Li
Collaborator: Dr. Zhipeng Wang
School of Life Sciences, Tsinghua
Teaching is the best way of learning. Together with Zhipeng Wang, a chemistry PhD student, I wrote parts of two introductory minireviews on chemical biology topics after serious literature research. These topics are NOT our specialties, but we got a better understanding of the fields through reading and writing the reviews.
Related publications:
- Wang, Z., Ding, X., Li, S., Shi, J., & Li, Y. (2014). Engineered fluorescence tags for in vivo protein labelling. RSC Advances. Link
- Wang, Z. P., Ding, X. Z., Wang, J., & Li, Y. M. (2015). Double-edged sword in cells: chemical biology studies of the vital role of cytochrome c in the intrinsic pre-apoptotic mitochondria leakage pathway. RSC Advances. Link
Screening for an improved genetically-encoded voltage indicator
2013 Summer
Advisor: Dr. Michael Lin (Lin lab)
Co-mentor: Dr. François St-Pierre (now St-Pierre lab)
Departments of Bioengineering and Pediatrics, Stanford
Brain processes information by electrical signals. It would be very helpful for us to understand how this happens if we can precisely record electrical activity in individual neurons or even whole neuronal circuits. Genetically encoded voltage indicators are tools that enable optical detection of the membrane potential of genetically defined neuronal circuits through fluorescence microscopy. At Stanford, I worked on a genetically encoded voltage indicator named Accelerated Sensor of Action Potentials (ASAP). ASAP is a fusion protein made of two modular components: 1) a membrane-localized voltage-sensitive domain and 2) an engineered GFP. The unique architecture of ASAP made its fluorescent intensity responsive to membrane voltage. Original ASAP sensors produced fast, millisecond kinetics, but was with limited fluorescence response to action potentials. In this project, I developed and validated a screening method to be used to search for improved sensors. I constructed 87 variants of the original sensor and screened them with this assay. Through the screening, I found a few promising variants with 50%-100% larger fluorescence responses.
The visit was sponsored by Stanford Undergraduate Visiting Researcher (UGVR) program.
Related publication:
- Yang, H. H.†, St-Pierre, F.†, Sun, X., Ding, X., Lin, M. Z., & Clandinin, T. R. (2016). Subcellular Imaging of Voltage and Calcium Signals Reveals Neural Processing In Vivo. Cell. †:Equal contribution. Link
Structural biology studies on a histone modification "reader"
2011, 2013 Fall
Advisor: Dr. Haitao Li (Dr. Haitao Li)
Co-mentor: Dr. Xiaonan Su
School of Medicine, Tsinghua
Histone post-translational modifications, or histone PTMs, play important roles in epigenetic regulation, and they are proposed to constitute a ‘histone code’. The role of a histone effector is to recognize the specific histone PTMs (or read the histone code) and recruit downstream proteins to take action. Previous reports showed that Spindlin-1 reads the trimethyl-lysine4 on histone H3 (H3K4me3). The structure of Spindlin-1 had already been solved before my project. However, the structure of free protein could not tell the molecular mechanism that Spindlin-1 recognizes the modification. The goal of this project was to solve the complex structure of Spindlin-1 bound to H3 peptide with trimethyl-lysine4 modification. We started from molecular cloning and expressing the protein in E.Coli, and established protocols for purifying the protein, and then screened for the crystallization conditions. After getting the co-crystal of the complex and solved the structure, I continued on biophysical characterization of the interaction with Isothermal Titration Calorimetry(ITC) and some mutant studies. Later structural studies showed a concurrent recognition of H3K4me3 and H3R8me2a by Spindlin-1.
This project marked the start of my research journey. The project was sponsored by National Training Programs of Innovation and Entrepreneurship for Undergraduate (China), and Tsinghua Talented Program ("Xuetangban" Program).Related publication:
Blog | 博客
2025年9月8日作为创业路上的小学生,今年勉强升到“二年级”,在这条走钢丝般的路上又有了一些新的观察和体会。总结为三点感悟,记录如下。 感悟一:关于“平台型 Biotech”的几个“常识”的二...从小到大,“考试”一直是我们所接受的教育的核心目标。而名校毕业生则是“考试”这类游戏中的佼佼者。然而,这些年来,走入现实世界中,我却发现,许多妨碍自己有效“做事”的思维习惯。恰恰是由十多...由于直接关系到人类健康,生物医药行业一方面追求创新、一方面也相当”保守“——这个行业受到高度监管、研发周期漫长,也因此很少看到年轻的创业者。甚至连博士毕业后直接创业的情况也并不多见。 不过近...(Translation by ChatGPT) 步骤 1. 阅读前人文献 (Du et al.) Step 1. Read the literature of...More Posts
Xiaozhe Ding © 2015
www.dingxiaozhe.com